We can perform both single- or paired-end sequencing up to 300bp read lengths.

NGS Submission Guidelines
Genomics Core Facility (GCF) – University of Bergen
Project meeting
We recommend to contact us and appoint a meeting to discuss the project, as early in the progress as possible. Ideally GCF and customer should have a meeting before any practical work has begun, to avoid typical pitfalls in project design. Especially when it comes to gene expression projects.
RNA-Sequencing
At the moment GCF offers several types of RNA-Seq protocols:
- TruSeq Stranded mRNA Library Prep protocol (sequencing polyA-transcripts from 3’end)
- TruSeq Stranded Total RNA Library Prep
- Whole transcriptome
- Depletion of ribosomal RNA
- NuGEN Universal Plus mRNA-Seq (Whole blood)
- PolyA-transcripts, but globin transcripts are depleted
Other protocols can be discussed, but a special agreement is then to be signed.
GCF strongly recommends the users to DNase treat their RNA samples before submitting them for sequencing
RNA QC:
Unless a quality check on total RNA is agreed with GCF in advance as a part of the service, please provide documentation on:
- Quantity
- A260/A280 and A260/A230 ratios (eg. NanoDrop)
- RNA integrity (eg. BioAnalyzer/TapeStation). GCF do not recommend to sequence RNA samples having a RIN > 7.
Normalize RNA:
All samples should be normalized (equal RNA concentration) before submission. For volume and concentration specifications, see table below.
RNA sample delivery to GCF
Please provide normalized RNA samples with a minimum amount of RNA, as specified in Table 1. RNA should be provided in a 96-well plate.
Please also provide a small aliquot (5-10ul) of normalized RNA to quality control, in a 96-well plate.
Considerations for RNA-Seq:
- How deep would you like to sequence your samples?
- mRNA sequencing or Whole transcriptome sequencing?
- Do your samples contain any highly abundant transcripts? Like RNA from blood, contains very high amounts of globin transcripts.
- Which organism is being sequenced? Some organisms that has less complex genomes needs to be sequenced together with a higher concentration of the artificial library, phiX.
The whole transcriptome protocol requires more reads per sample, and hence is a more expensive alternative. But gives you the option of more novel discoveries such as non-coding RNA, fusion transcripts and alternative isoforms.
RNA project design
Please contact GCF on beforehand to get help on project design. It is important that different treatments/study groups are balanced in the experiment and RNA extraction procedure. A non-balanced design could lead to technical batch effects in the data.
Whole Genome Sequencing
At the moment GCF offers two standard protocols for whole genome sequencing:
- Illumina TruSeq PCR-free protocol ( > 1ug genomic DNA available).
- Illumina TruSeq Nano protocol (100ng genomic DNA as input, includes an amplification step)
Other protocols can be discussed, but a special agreement is then to be signed.
Unless a quality check on genomic DNA is agreed with GCF in advance as a part of the service, please provide agarose gel pictures, TapeStation analysis or other kind of documentation.
All samples should be normalized (equal DNA concentration) before submission. For volume and concentration specifications, see table below.
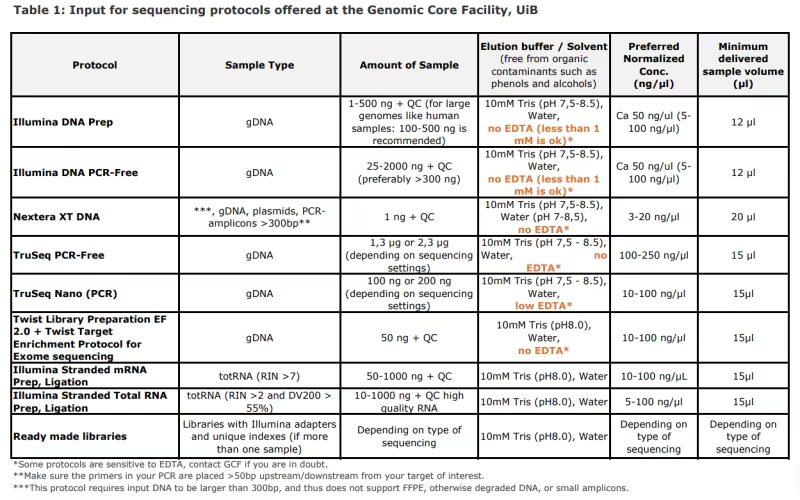
Table 1: Input for sequencing protocols offered at the Genomic Core Facility, UiB